research
My work centers on bringing statistical insight and understanding to the practice of data science. As a classically-trained statistician with an extensive track record of collaborative work, industry experience, as well as experience with modern machine learning methods, I seek to identify the challenges that arise from contemporary data-driven inquiry, and to develop statistical frameworks to solve them.
Data science for generative AI:
During my postdoctoral training, I am particularly enthusiastic about exploring the data science inquires arising from advanced AI techniques, such as large language models (LLMs) and text-to-image models (e.g., DALLE and Stable Diffusion).
One line of my work applies large language models (LLMs) to single-cell genomics: While there has been significant recent progress in foundation models for single-cell data, existing models require extensive data curation and resource-intensive training on tens of millions of cells. We explored a much simpler alternative, GenePT, by leveraging LLMs embeddings of genes and cells based on descriptions of their functionalities in the literature. On many downstream tasks, GenePT achieves comparable, and often better, performance than existing single-cell foundation models.
I have also introduced TWIGMA, a comprehensive dataset of generative-AI images on Twitter. We found that gen-AI images possess distinctive characteristics and exhibit, on average, lower variability when compared to their non-gen-AI counterparts. Additionally, we characterize the longitudinal shift in the themes of AI-generated images by users.
Selected Manuscripts:
- Chen YT, Zou J (2023+). A Simple But Hard-to-Beat Foundation Model for Genes and Cells Built From ChatGPT.
- Preprint: https://www.biorxiv.org/content/10.1101/2023.10.16.562533v1
- A preliminary of this work has been accepted as an oral presentation at MLCB 2023 (< 15% acceptance rate).
- Chen YT, Zou J (2023). TWIGMA: A dataset of AI-Generated Images with Metadata From Twitter. NeurIPS 2023. arXiv link: https: //arxiv.org/abs/2306.08310.
Testing data-driven hypotheses
In my PhD research, I tackled the problem of testing data-driven hypotheses generated by unsupervised learning methods such as changepoint detection and clustering. For instance, in single-cell RNA-sequencing analyses, researchers often first cluster the cells, and then test for a difference in the expected gene expression levels between the clusters. However, this is invalid from a statistical perspective: once we have used the data to generate hypotheses, standard statistical inference tools will no longer be valid (e.g., an extremely inflated Type I error rate). I have developed valid inference approaches in this setting, and have applied them to data from genomics, neuroscience, and epidemiology.
Selected Manuscripts:
-
Clustering And Differential Expression Testing: Chen YT, Gao LL (2023+). Testing for a difference in means of a single feature after clustering. arXiv link: https://arxiv.org/abs/2311.16375.
-
Inference after k-means clustering: Chen YT, Witten DM (2023). Selective inference for k-means clustering. Journal of Machine learning Research. arXiv link: https://arxiv.org/abs/2203.15267.
-
Inference after graph fused lasso: Chen YT, Jewell SW, and Witten DM (2023). Journal of Computational and Graphical Statistics. arXiv link: https://arxiv.org/abs/2109.10451.
-
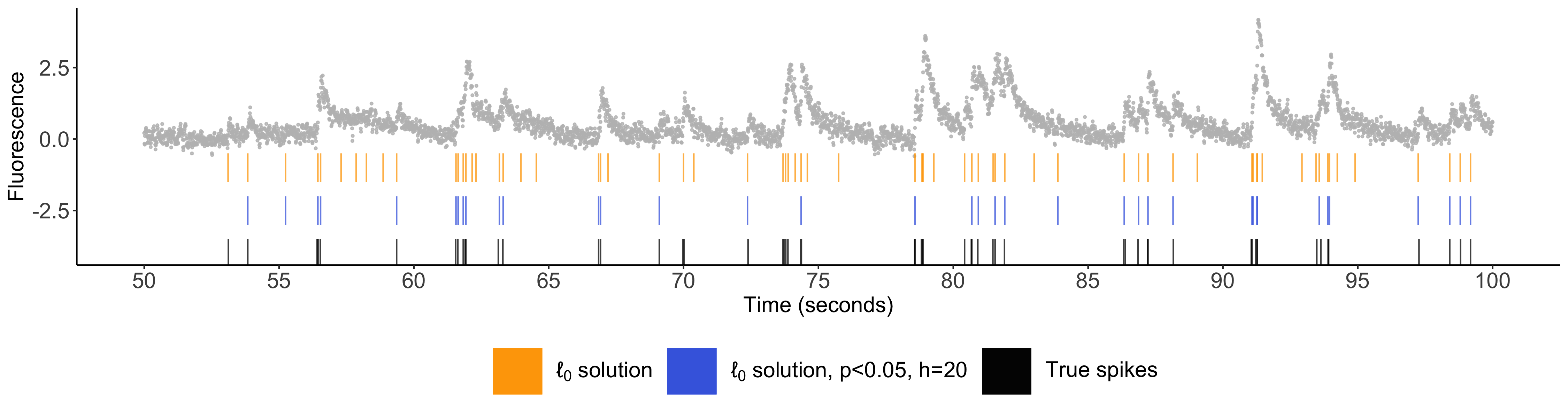
Inference after spike detection: Chen YT, Jewell SW, Witten DM. (2022) Quantifying uncertainty in spikes estimated from calcium imaging data, Biostatistics, Oxford University Press.
Selected Software:
- CADET: https://yiqunchen.github.io/CADET/
- KmeansInference: https://yiqunchen.github.io/KmeansInference/
- GFLassoInference: https://yiqunchen.github.io/GFLassoInference/
- SpikeInference: https://github.com/yiqunchen/SpikeInference
Selected Talks:
-
You can learn more about my work on “selective inference for k-means clustering” at International Seminar on Selective Inference here
-
You can learn more about my work on “Quantifying uncertainty in spikes estimated from calcium imaging data” at WNAR 2021 here
Selected applications to (biomedical) data science:
Race and Ethnicity in Research Data Collection:
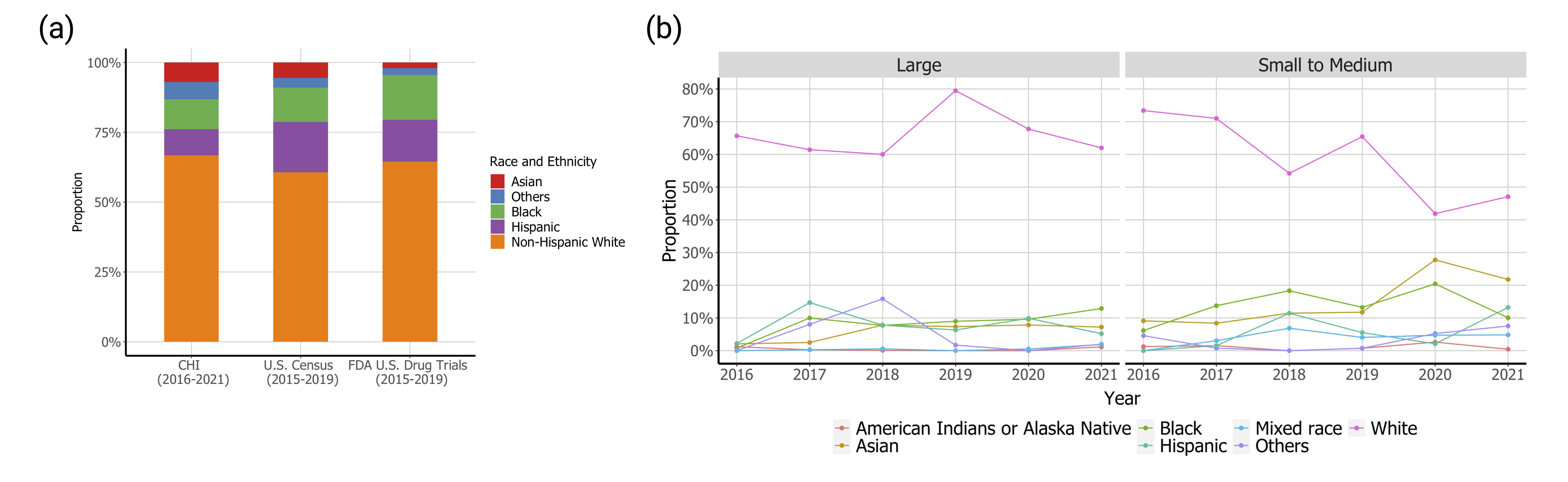
In today’s data-driven era, statisticians should play an active role in study design and data collection. Collaborating with researchers from human-computer interaction (HCI) - a field crucial for spawning innovative technologies and interfaces — and computing ethics, I investigated the racial and ethnic diversity of research participants in Human-Computer Interaction (HCI). Our work spotlighted a conspicuous absence of the documentation of participants’ racial and ethnic backgrounds. Together with colleagues, I consequently formulated guidelines for HCI researchers regarding the collection of race and ethnicity data, and emphasized the pressing need for conscious decision-making rather than passive omissions, ensuring the creation of truly inclusive and equitable technologies.
Selected Publications:
- Chen YT, Smith AD, Reinecke K, and To A (2023). Why, when, and from whom: considerations for collecting and reporting race and ethnicity data in HCI. In CHI’23.
- Awarded Best Paper Honorable Mention (Top 5% of all submissions to CHI 2023); also covered in https://www.khoury.northeastern.edu/awards-ethics-cross-college-collaboration-northeastern-at-chi-2023.
- This paper reviewed six years of CHI proceedings and interviewed 15 authors to understand the current practice of collecting race and ethnicity data in the field of Human-computer Interaction. It highlights important considerations for researchers when they decide to collect such data.
- Chen YT, Smith AD, Reinecke K, and To A (2022). Collecting and Reporting Race and Ethnicity Data in HCI. In CHI’22 Extended Abstracts.
Social networks characteristics and HIV/TB care outcomes in sub-Saharan Africa
Social isolation among HIV-positive persons might be an important barrier to care. Similarly, analyzing one’s social network could elucidate Tuberculosis (TB) transmission dynamics outside of their home and may inform novel, network-based case-finding strategies.
We used data collected as a part of the SEARCH Study in rural Kenya and Uganda, and constructed 32 community-wide, sociocentric networks. We found that (i)HIV-positive persons named as a contact by fewer people may be at higher risk for poor HIV care outcomes,suggesting opportunities for targeted interventions; and (ii) social networks with higher centrality, more men, contacts with HIV, and TB infection, were positively associated with TB infection.

Selected Publications:
-
TB infections and social networks: Chen YT†, Marquez C†, Atukunda M, Chamie G, Balzer LB, …, Charlebois ED, Havlir DV, Petersen ML (2022+). The Association Between Social Network Characteristics and TB Infection Among Adults in Nine Rural Ugandan Communities. To appear in Clinical Infectious Diseases; † denotes joint first authorship.
-
HIV care outcome and social networks: Chen YT, Brown LB, Chamie G, Kwarisiima D, Ayieko J, Kabami J, Charlebois E, Clark T, Kamya M, Havlir DV, Petersen ML, and Balzer LB (2021). Social networks and HIV care outcomes in rural Kenya and Uganda. Epidemiology, 32(4):551-559.
-
Two posters at CROI 2020:
Software engineering:
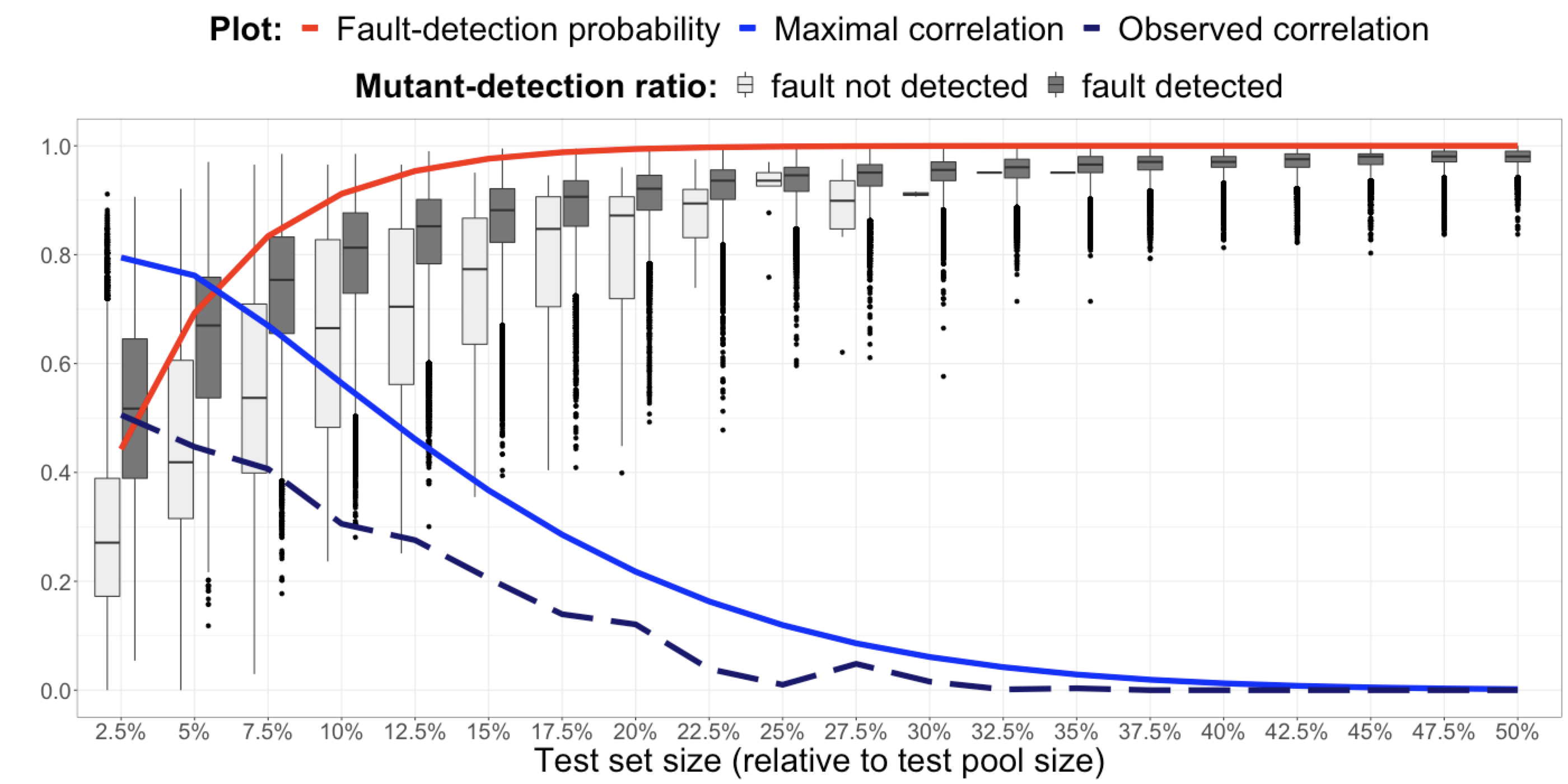
The relationship between fault detection, test adequacy criteria, and test set size remains a complex yet central topic in software engineering research. In this project, we addressed the supposed contradiction of prior work and explains why test set size is neither a confounding variable, as previously suggested, nor an independent variable that should be experimentally manipulated. An alternative methodology is proposed for comparing test adequacy criteria on an equal basis, which accounts for test set size without directly manipulating it through unrealistic stratification.
Manuscript:
- Chen YT, Gopinath R, Tadakamalla A, Ernst MD, Holmes R, Fraser G, Am- mann P, Just R. Revisiting the relationship between fault detection, test adequacy criteria, and test set size. In: 2020 35th IEEE/ACM International Conference on Automated Software Engineering (ASE). 2020:237-249.
- Link to the publisher’s version here.
- You can learn more about this project by watching my talk at ASE 2020 here