What is KmeansInference?
KmeansInference is an R package for testing for a difference in means between clusters of observations identified via k-means clustering.
How do I install the package?
To download (and load) the KmeansInference package, use the code below.
require("devtools")
devtools::install_github("yiqunchen/KmeansInference")
library(KmeansInference)Why do we need KmeansInference?
Testing for a difference in means between two groups is one of the most fundamental tasks in statistics, with numerous applications. If the groups under investigation are pre-specified, i.e., defined without making use of the observed data, then classical hypothesis tests will control the Type I error rate.
However, in practice, data analysts often find themselves testing the null hypothesis of a difference in means between two groups that are defined through the same data used for testing, e.g., via the output of a clustering algorithm. For instance, in single-cell RNA-sequencing analysis, researchers often first cluster the cells, and then test for a difference in the mean gene expression levels between the clusters to quantify up- or down-regulation of genes and identify new cell types. In this case, because the groups are defined through the data, classical hypothesis tests lead to an inflated Type I error.
In particular, the inferential challenges resulting from testing data-guided hypotheses have been described as a “grand challenge” in the field of genomics (Lähnemann et al., 2020), and papers in the field continue to overlook this issue: as an example, seurat (Stuart et al., 2019), the state-of-the-art single-cell RNA sequencing analysis tool, tests for differential gene expression between groups obtained via clustering, with a note that “p-values [from these hypotheses] should be interpreted cautiously, as the genes used for clustering are the same genes tested for differential expression.”
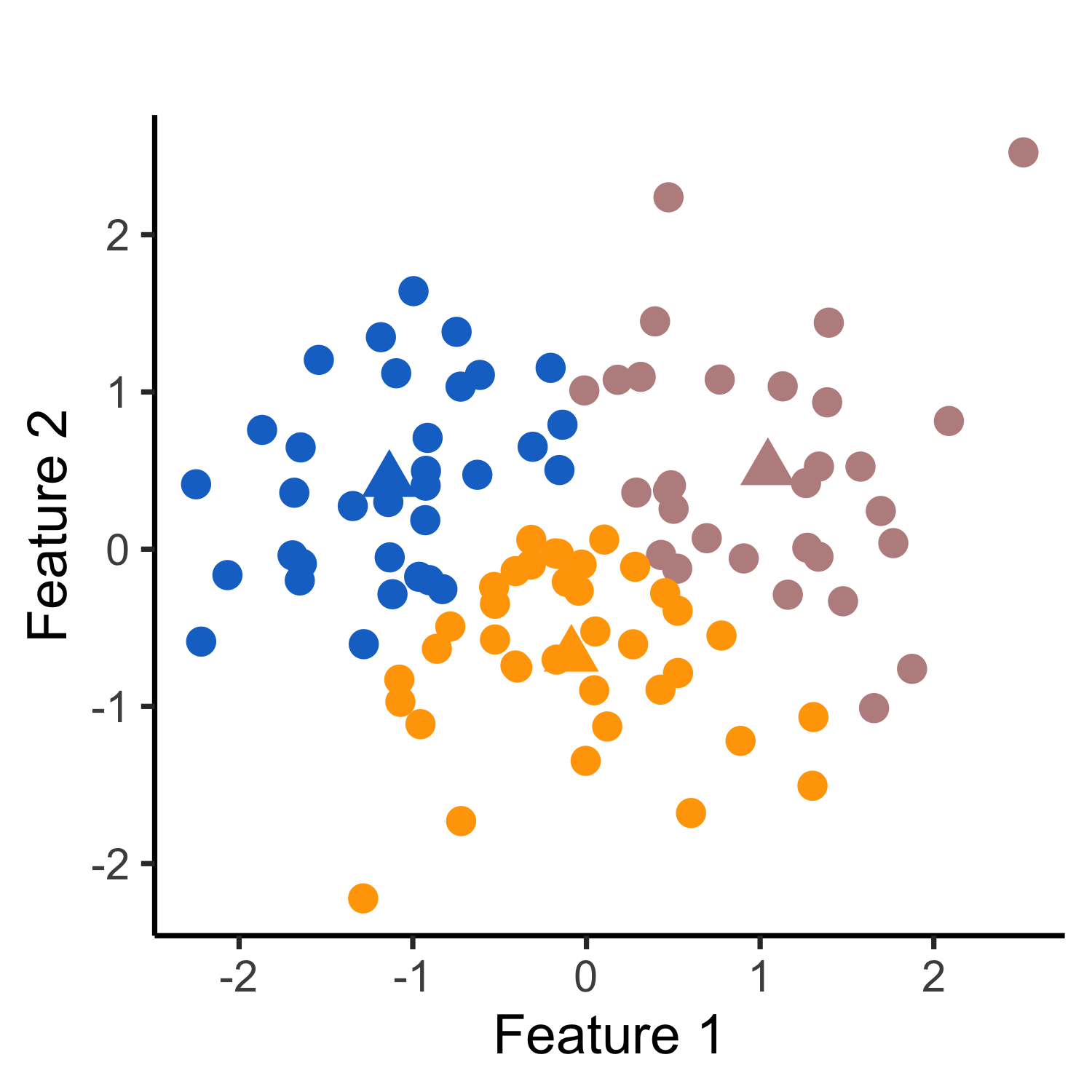
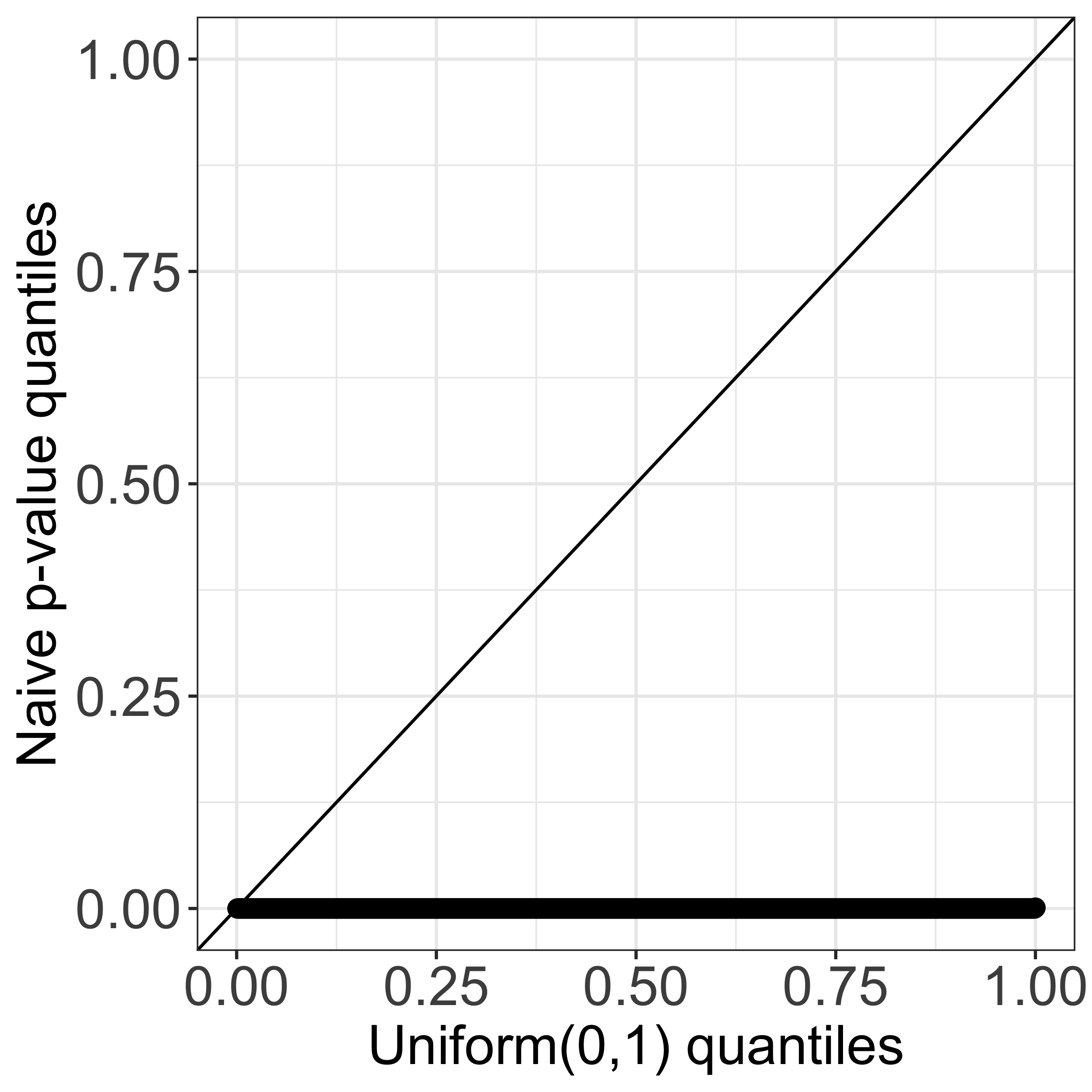
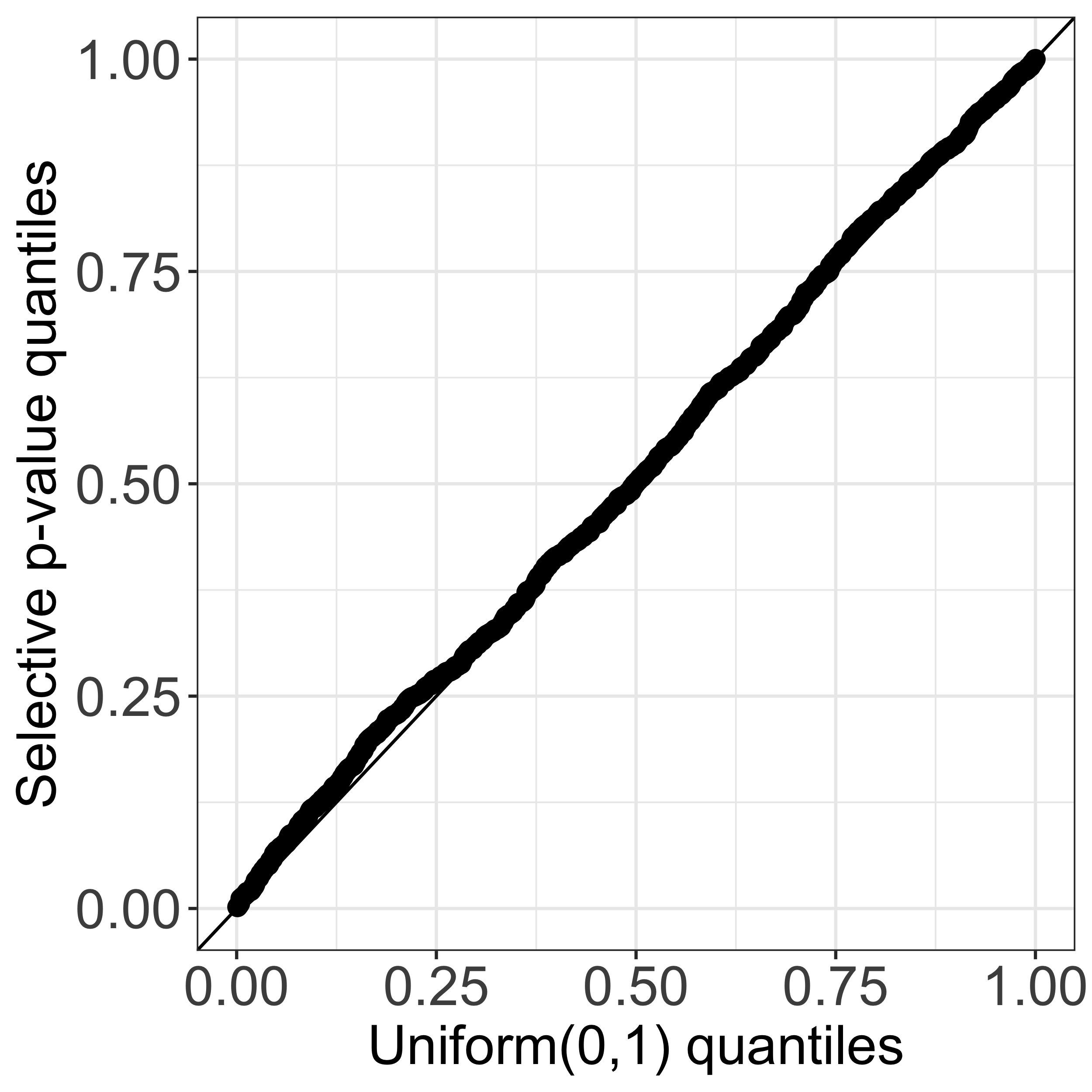
In this work, we propose a test for a difference in means between two clusters estimated from applying k-means clustering. Given a set of observations, suppose that we first apply k-means clustering to obtain clusters. We might then consider testing the null hypothesis that the mean is the same across two estimated clusters. As an example, consider a simulated data set where x1, …, x150 ∼ 𝒩(02,I2), and applying k-means clustering with K=3 yields three clusters (see leftmost panel of Figure 1). In the middle panel, we display the quantiles of the naive p-value for testing the null hypothesis that a randomly chosen pair of estimated clusters have the same mean. Because the naive p-value does not account for the fact that the cluster labels are obtained through the same data used for testing, the corresponding test leads to an extremely inflated Type I error. By contrast, our selective p-value, conditional on the event that k-means clustering yields the two estimated clusters under investigation, leads to a correctly-sized test (see the rightmost panel of Figure 1). More detailed simulation results can be found in Section 2 of our manuscript.
Link to additional resources
- You can learn more about the technical details in our manuscript and in the technical details section.
- You can learn more about how to use our software in the tutorials section.
- Finally, code and steps to reproduce the figures in our manuscript can be found in the GitHub repo https://github.com/yiqunchen/KmeansInference-experiments.
Citation
If you use KmeansInference for your analysis, please cite our manuscript:
Chen YT, Witten DM. (2022+) Selective inference for k-means clustering. arXiv preprint. https://arxiv.org/abs/2203.15267.
Bug Reports / Change Requests
If you encounter a bug or would like to make a change request, please file it as an issue here.
References
Chen YT, Witten DM. (2022+) Selective inference for k-means clustering. arXiv preprint. https://arxiv.org/abs/2203.15267.
Fithian W, Sun D, Taylor J. (2014) Optimal Inference After Model Selection. arXiv:1410.2597 [mathST].
Gao, L. L., Bien, J., and Witten, D. (2022+). Selective inference for hierarchical clustering. arXiv:2012.02936.
Lähnemann, D., Köster, J., Szczurek, E., et al. (2020). Eleven grand challenges in single-cell data science. Genome Biology, 21(1):31.
Stuart, T., Butler, A., Hoffman, P., et al. (2019). Comprehensive integration of Single-Cell data. Cell, 177(7):1888–1902.e21.