Test for a difference in means between clusters of observations identified via k-means clustering.
kmeans_inference.RdThis function tests the null hypothesis of no difference in means between
output by k-means clustering. The clusters are numbered as per the results of
the kmeans_estimation function in the KmeansInference package.
kmeans_inference(
X,
k,
cluster_1,
cluster_2,
iso = TRUE,
sig = NULL,
SigInv = NULL,
iter.max = 10,
seed = 1234,
tol_eps = 1e-06,
verbose = TRUE
)Arguments
- X
Numeric matrix; \(n\) by \(q\) matrix of observed data
- k
Integer; the number of clusters for k-means clustering
- cluster_1, cluster_2
Two different integers in 1,...,k; two estimated clusters to test, as indexed by the results of
kmeans_estimation.- iso
Boolean. If TRUE, an isotropic covariance matrix model is used.
- sig
Numeric; noise standard deviation for the observed data, a non-negative number; relevant if
iso=TRUE. If it's not given as input, a median-based estimator will be by default (see Section 4.2 of our manuscript).- SigInv
Numeric matrix; if
isois FALSE, *required* \(q\) by \(q\) matrix specifying \(\Sigma^{-1}\).- iter.max
Positive integer; the maximum number of iterations allowed in k-means clustering algorithm. Default to
10.- seed
Random seed for the initialization in k-means clustering algorithm.
- tol_eps
A small number specifying the convergence criterion for k-means clustering, default to
1e-6.
Value
Returns a list with the following elements:
p_naivethe naive p-value which ignores the fact that the clusters under consideration are estimated from the same data used for testingpvalthe selective p-value \(p_{selective}\) in Chen and Witten (2022+)final_intervalthe conditioning set of Chen and Witten (2022+), stored as theIntervalsclasstest_stattest statistic: the difference in the empirical means of two estimated clustersfinal_clusterEstimated clusters via k-means clustering
Details
For better rendering of the equations, visit https://yiqunchen.github.io/KmeansInference/reference/index.html.
Consider the generative model \(X ~ MN(\mu,I_n,\sigma^2 I_q)\). First recall that k-means clustering
solves the following optimization problem
$$ \sum_{k=1}^K \sum_{i \in C_k} \big\Vert x_i - \sum_{i \in C_k} x_i/|C_k| \big\Vert_2^2 , $$
where \(C_1,..., C_K\) forms a partition of the integers \(1,..., n\), and can be regarded as
the estimated clusters of the original observations. Lloyd's algorithm is an iterative apparoach to solve
this optimization problem.
Now suppose we want to test whether the means of two estimated clusters cluster_1 and cluster_2
are equal; or equivalently, the null hypothesis of the form \(H_{0}: \mu^T \nu = 0_q\) versus
\(H_{1}: \mu^T \nu \neq 0_q\) for suitably chosen \(\nu\) and all-zero vector \(0_q\).
This function computes the following p-value: $$P \Big( || X^T\nu || \ge || x^T\nu ||_2 \; | \; \bigcap_{t=1}^{T}\bigcap_{i=1}^{n} \{ c_i^{(t)}(X) = c_i^{(t)}( x ) \}, \Pi Y = \Pi y \Big),$$ where \(c_i^{(t)}\) is the is the cluster to which the \(i\)th observation is assigned during the \(t\)th iteration of the Lloyd's algorithm, and \(\Pi\) is the orthogonal projection to the orthogonal complement of \(\nu\). In particular, the test based on this p-value controls the selective Type I error and has substantial power. Readers can refer to the Sections 2 and 4 in Chen and Witten (2022+) for more details.
References
Chen YT, Witten DM. (2022+) Selective inference for k-means clustering. arXiv preprint. https://arxiv.org/abs/2203.15267. Lloyd, S. P. (1957, 1982). Least squares quantization in PCM. Technical Note, Bell Laboratories. Published in 1982 in IEEE Transactions on Information Theory, 28, 128–137.
Examples
library(KmeansInference)
library(ggplot2)
set.seed(2022)
n <- 150
true_clusters <- c(rep(1, 50), rep(2, 50), rep(3, 50))
delta <- 10
q <- 2
mu <- rbind(c(delta/2,rep(0,q-1)),
c(rep(0,q-1), sqrt(3)*delta/2),
c(-delta/2,rep(0,q-1)) )
sig <- 1
# Generate a matrix normal sample
X <- matrix(rnorm(n*q, sd=sig), n, q) + mu[true_clusters, ]
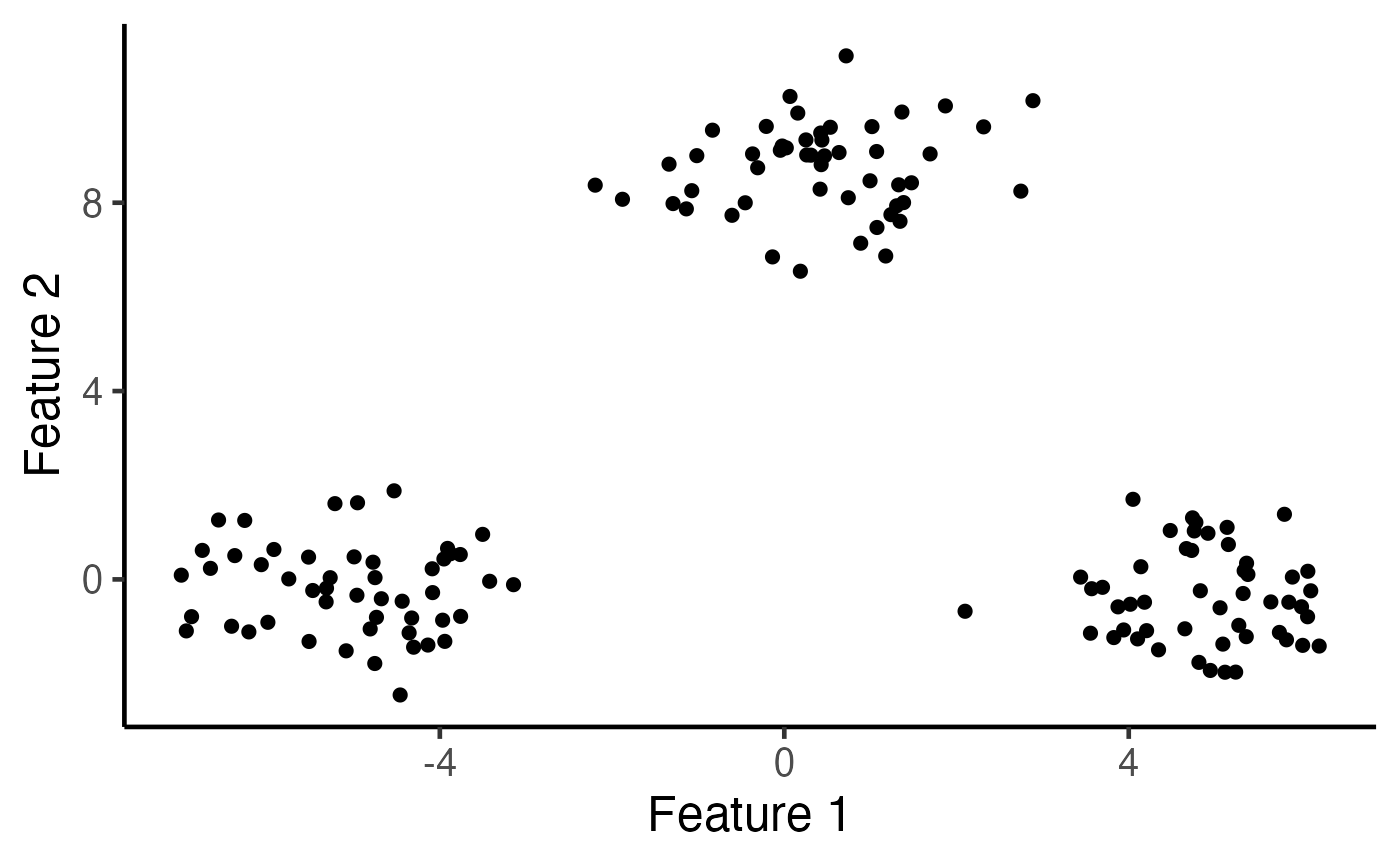
# Visualize the data
ggplot(data.frame(X), aes(x=X1, y=X2)) +
geom_point(cex=2) + xlab("Feature 1") + ylab("Feature 2") +
theme_classic(base_size=18) + theme(legend.position="none") +
scale_colour_manual(values=c("dodgerblue3", "rosybrown", "orange")) +
theme(legend.title = element_blank(),
plot.title = element_text(hjust = 0.5))
 k <- 3
# Run k-means clustering with K=3
estimated_clusters <- kmeans_estimation(X, k,iter.max = 20,seed = 2021)$final_cluster
table(true_clusters,estimated_clusters)
#> estimated_clusters
#> true_clusters 1 2 3
#> 1 0 50 0
#> 2 0 0 50
#> 3 50 0 0
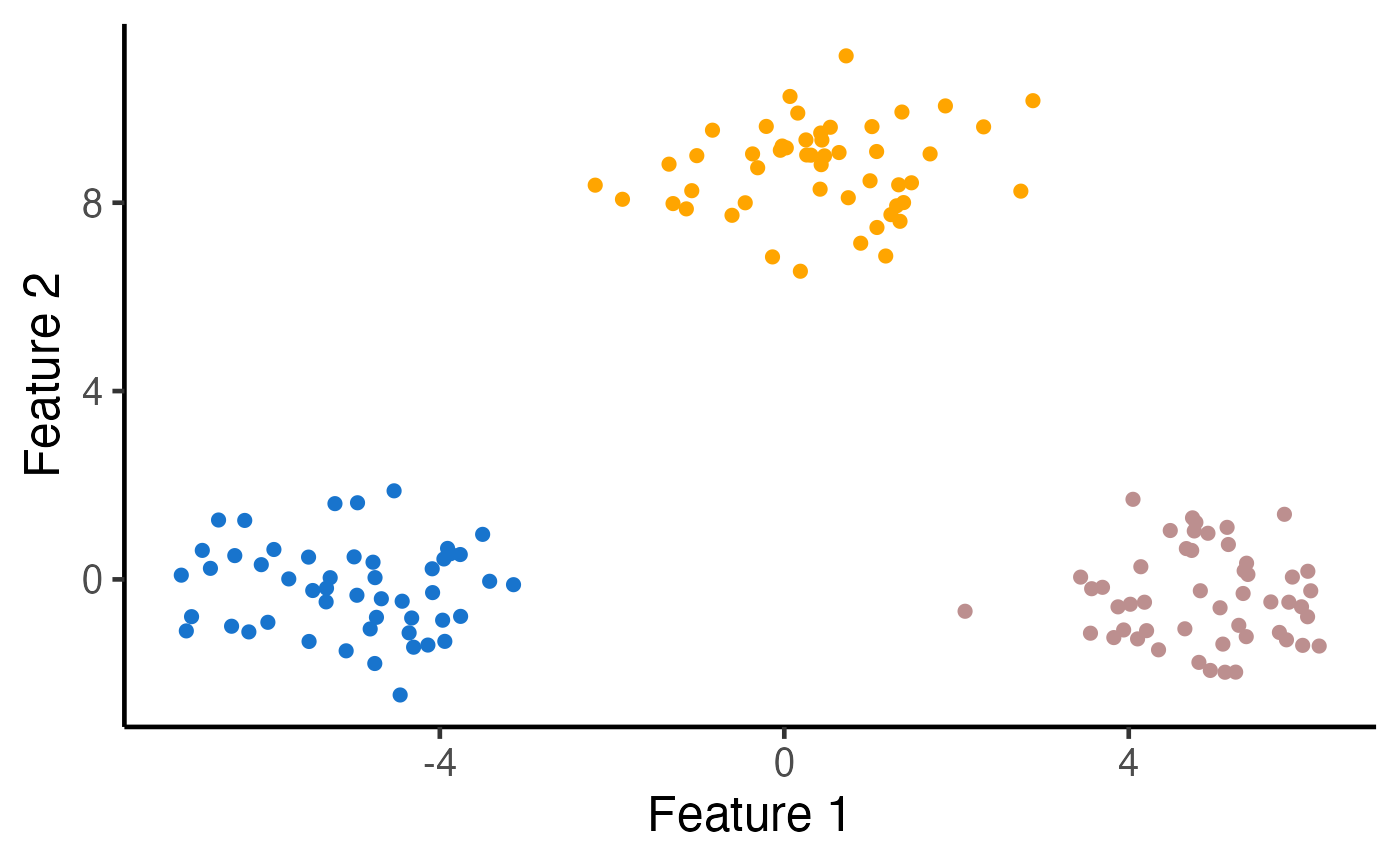
# Visualize the clusters
ggplot(data.frame(X), aes(x=X1, y=X2, col=as.factor(estimated_clusters))) +
geom_point(cex=2) + xlab("Feature 1") + ylab("Feature 2") +
theme_classic(base_size=18) + theme(legend.position="none") +
scale_colour_manual(values=c("dodgerblue3", "rosybrown", "orange")) +
theme(legend.title = element_blank(), plot.title = element_text(hjust = 0.5))
k <- 3
# Run k-means clustering with K=3
estimated_clusters <- kmeans_estimation(X, k,iter.max = 20,seed = 2021)$final_cluster
table(true_clusters,estimated_clusters)
#> estimated_clusters
#> true_clusters 1 2 3
#> 1 0 50 0
#> 2 0 0 50
#> 3 50 0 0
# Visualize the clusters
ggplot(data.frame(X), aes(x=X1, y=X2, col=as.factor(estimated_clusters))) +
geom_point(cex=2) + xlab("Feature 1") + ylab("Feature 2") +
theme_classic(base_size=18) + theme(legend.position="none") +
scale_colour_manual(values=c("dodgerblue3", "rosybrown", "orange")) +
theme(legend.title = element_blank(), plot.title = element_text(hjust = 0.5))
 ### Run a test for a difference in means between estimated clusters 1 and 3
cluster_1 <- 1
cluster_2 <- 3
cl_1_2_inference_demo <- kmeans_inference(X, k=3, cluster_1, cluster_2,
sig=sig, iter.max = 20, seed = 2021)
summary(cl_1_2_inference_demo)
#> cluster_1 cluster_2 test_stat p_selective p_naive
#> 1 1 3 10.44088 4.473563e-15 0
### Run a test for a difference in means between estimated clusters 1 and 3
cluster_1 <- 1
cluster_2 <- 3
cl_1_2_inference_demo <- kmeans_inference(X, k=3, cluster_1, cluster_2,
sig=sig, iter.max = 20, seed = 2021)
summary(cl_1_2_inference_demo)
#> cluster_1 cluster_2 test_stat p_selective p_naive
#> 1 1 3 10.44088 4.473563e-15 0